Contents
SynTouch Cheetah Package
This page is currently under development, please check back in the future for updates
Welcome to SynTouch's new ROS package st_cheetah. SynTouch is currently developing this package to collect BioTac data from the Cheetah SPI/USB controller. Collected data can be processed using the package st_biotac.
For more information visit: SynTouch Website
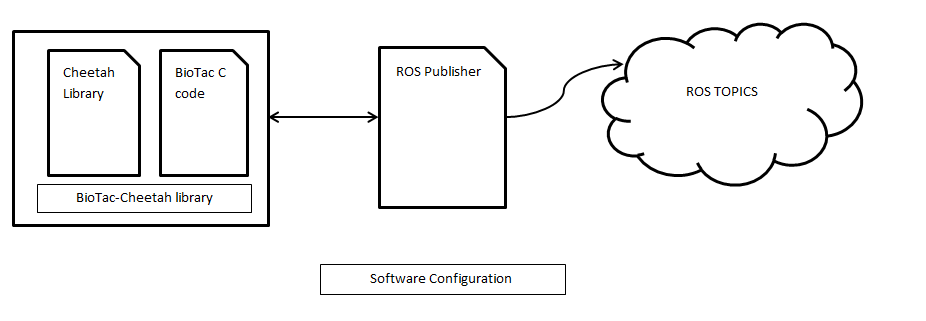
st_cheetah
This package contains ROS Drivers for collecting BioTac sensor data from the Cheetah SPI Host controller (Total Phase) and multi-BioTac board. The contained node publishes raw data from the attached BioTacs and writes their properties to the parameter server.
Authors: Abhishek Pawar, Gary Lin, and Jeremy Fishel, contact: ros@syntouchllc.com
Maintained by: Abhishek Pawar / abhishek.pawar@syntouchllc.com
- License: GPL
External Website: http://www.syntouchllc.com
- Source: (software currently under development)
Overview
This package provides a ROS interface for the !BioTac sensors through the Cheetah SPI Port. The Cheetah device samples the connected !BioTacs according to the specified sampling parameters. The !BioTac C library is included in this package to communicate with the sensors and decode the data. Each sample is then published as a new message that contains the channel identifier, sample data for each attached !BioTac, and a timestamp.

NOTE: Do not Download the latest library from Total Phase. Our software is only compatible with the version that is included in this package.
BioTac Communication and Batch Structure
One or more BioTac sensors can be connected to the multi-BioTac board and Cheetah SPI Host which is in turn connected to the computer via a USB cable.
The Cheetah SPI Host is capable of queueing batches of BioTac SPI sampling commands that execute asynchronously and are transmitted over USB to the host computer when they become available. The BioTac C Library takes advantage of these asynchronous shifts to ensure continuous sampling of BioTac data even if USB communication is delayed. During initialization, 16 batches of data are shifted to the host adapter's queue as a buffer to ensure continuously sampled data. Each time a batch is collected an identical batch of sampling commands is added at the end of the queue. For this reason the minimum batch size is one complete sampling sequence, or a frame. An integral multiple of frames can be structured into a batch. Using more frames per batch is efficient and reduces the USB bandwidth of the computer, which may be necessary for slower computers; fewer frames per batch allows for faster access to data, but may cause discontinuous sampling if the computer is not fast enough to keep up. When this happens and the computer cannot shift batches to the Cheetah SPI host faster than they are acquired it is referred to as buffer underrun and indicates that the collected BioTac data is not continuous. When this happens, increasing the batch length can solve the problem. Setting this value generally requires some tuning, it is recommended that you start with a batch length of 30ms and adjust either higher or lower depending on computer performance. Program behavior in response to buffer underrun can be configured as indicated below.
ROS API
Rosgraph containing the nodes and topics
BioTac_Cheetah_Pub
This node finds all available Cheetah devices and references the configuration file to determine which device to communicate with (specified by serial number). Then it checks for available BioTacs and retrieves their device information. After this configuration is complete it begins sampling and publishing a continuous stream of data.Published Topics
/biotacs/<port_name> (st_biotac/BioTac_Port_Data)- This topic contains BioTac Data from all BioTacs connected to the Cheetah port. The name of the topic depends on the parameter (~cheetah_port_name) as specified in the configuration file (default value is PORT0). For data structure refer to BioTac Messages.
Parameters
Cheetah Hardware Configuration parameters

~cheetah_serial_num (string, default: "0")
- Serial number of the Cheetah device connected to the BioTacs (with dashes). It can be found on the cheetah device as shown in the image above. If this value is set to 0 the first available Cheetah device is used. If this value is set to the particular device connected then connection to that device is established if it is found. For systems using more than one Cheetah this must be configured to avoid problems.
- The identifier of the cheetah device(i.e. left_hand). This parameter is used to specify the topic associated with this Cheetah.
Sampling Parameters
~bt_sample_rate_hz (int, default: 4400)
- Sampling rate of the BioTacs in Hz (maximum recommended rate is 20kHz). This is the overall sampling rate, sampling rate of each channel depends on frame structure (for the default frame structure, PAC sampling rate is 1/2 of the overall sampling rate, and all other channels are sampled at 1/44 of the overall sampling rate).
- The sampling pattern for the BioTac. They are hard coded in the program and can be identified by numbers as listed below:
0 - Default Sampling Sequence - Vibrations (PAC) signals are sampled at high frequency and interleaved with all other measurements. This is the recommended sampling sequence for most applications and is recommended to have an overall sampling rate of 4400Hz.
Sequence (44 Samples/frame): PAC, E1, PAC, E2, ... PAC, E18, PAC, E19, PAC, PDC, PAC, TAC, PAC, TDC,
1 - Default with Hall - This sampling sequence is for BioTacs versions 2.3 and later with integrated Hall Sensor that require the sensing of this channel. The recommended sampling rate for this sequence is 4600Hz.
Sequence (46 Samples/frame): PAC, E1, PAC, E2, ... PAC, E18, PAC, E19, PAC, PDC, PAC, TAC, PAC, TDC, PAC, Hall
2 - All Channels - This sampling sequence is recommended when high bandwidth vibration data is not necessary. It reduces the sampling rate of PAC to conserve bandwidth. Recommended sampling rate is 2400Hz, however lower sampling rates can be used if desired. NOTE: when using this sampling sequence PAC is vulnerable to aliasing.
Sequence (24 Samples/frame): PAC, PDC, TAC, TDC, Hall, E1, E2, ... E18, E19,
3 - Electrodes Only - This sampling sequence is recommended for force extraction algorithms that make use of the BioTac electrodes, preferably for light forces. The recommended sampling rate is 1900Hz.
Sequence (19 Samples/frame): E1, E2, ... E18, E19,
4 - AC/DC Pressure - This sampling sequence is recommended for high-resolution sampling of vibration signals and contact, it alternates between the PAC and PDC channels. The recommended sampling rate is 4400Hz.
Sequence (2 Samples/frame): PAC, PDC,
5 - AC Pressure Only - This sampling sequence is recommended for extremely low-noise vibration measurements. The recommended sampling rate is 2200Hz.
Sequence (2 Samples/frame): PAC, PDC,
Batch Parameters
~bt_batch_ms (int, default: 30)
- The Cheetah SPI device asynchronously collects data and transmits multiple frames of data in batches over USB protocol to the host computer. Shorter batches allow for faster response by the host but can result in excessive USB traffic. In slower computers, this can result in breaks in the asynchronous sampling. If these problems are encountered, increasing the batch length (so more frames are transmitted in each batch communication) usually fixes the problem. The recommended batch length for most systems is 30ms.
- This parameter specifies how the BioTac_Cheetah_Pub node will behave if the buffer in the Cheetah has underrun. The following parameters can be set:
- 0 - display warning and continue sampling
- 1 - display warning and reset sampling sequence before continuing
2 - Display error and terminate program (recommended for time-critical applications)
For more details go here.
BioTac Properties
~bt_status (bool[], default: NULL)
- An array of boolean variables that indicate which connectors have BioTacs attached (1=connected, 0=disconnected).
- An array containing the serial numbers of attached BioTacs.
- An array containing the flex circuit version of attached BioTacs.
- An array containing the firmware version of attached BioTacs.
- An array containing the CPU speed of attached BioTacs in MHz.
Other Parameteres
~print_to_console (int, default: 0)
- Flag to print the debug output and samples to the screen (0 = do not print, 1 = print).
Documentation
The BioTac_Cheetah_Pub node performs a number of error checking operations to ensure hardware is properly configured and newly attached or disconnected BioTacs are identified. On each execution the node does the following:
- Checks if the specified Cheetah device is connected to the host computer and displays warnings if not.
Checks if any BioTacs are connected and displays warnings if there are none. The BioTac information for each attached BioTac is stored in global parameters.
- The publisher configures the Cheetah for asynchronous sampling.
- The publisher collects a batch from cheetah and publishes samples.
If a BioTac is disconnected or reconnected it updates the parameters. If a new BioTac is connected sampling is stopped to query the BioTac properties before restarting sampling.
Installation Instructions
Download the SynTouch stack from (link). Unpack the package in a working ROS directory.
$ rosmake st_cheetah
Configure the launch file parameters. The minimum configuration is specified below:
<node name="Biotac_Cheetah_Pub" pkg="st_cheetah" type="Biotac_Cheetah_Pub"> <param name="cheetah_serial_num" type="string" value="0"> <param name="cheetah_port_name" type="string" value="Port0"> </node>
Or the extended configuration:
<node name="Biotac_Cheetah_Pub" pkg="st_cheetah" type="Biotac_Cheetah_Pub"> <param name="cheetah_serial_num" type="string" value="0"> <param name="cheetah_port_name" type="string" value="Port0"> <param name="bt_sample_rate_hz" type="int" value=4400> <param name="bt_frame_type" type="int" value=0> <param name="bt_batch_ms" type="int" value=30> <param name="bt_buffer_underrun_option" type="int" value=0> <param name="print_to_console" type="int" value=0> </node>
Run the launch file with:
$ roslaunch st_cheetah st_cheetah.launch
Contributing
If you are interested in contributing to the development of this stack or would like to add or help us add new function to a package please contact us at ros@syntouchllc.com
Support
If you encounter a problem or need support please contact us at ros@syntouchllc.com







